Abstract
Soybean is highly sensitive to flooding and extreme rainfall. The phenotypic variation of flooding tolerance is a complex quantitative trait controlled by many genes and their interaction with environmental factors. We previously constructed a gene-pool relevant to soybean flooding-tolerant responses from integrated multiple omics and non-omics databases, and selected 144 prioritized flooding tolerance genes (FTgenes). In this study, we proposed a comprehensive framework at the systems level, using competitive (hypergeometric test) and self-contained (sum-statistic, sum-square-statistic) pathway-based approaches to identify biologically enriched pathways through evaluating the joint effects of the FTgenes within annotated pathways. These FTgenes were significantly enriched in 36 pathways in the Gene Ontology database. These pathways were related to plant hormones, defense-related, primary metabolic process, and system development pathways, which plays key roles in soybean flooding-induced responses. We further identified nine key FTgenes from important subnetworks extracted from several gene networks of enriched pathways. The nine key FTgenes were significantly expressed in soybean root under flooding stress in a qRT-PCR analysis. We demonstrated that this systems biology framework is promising to uncover important key genes underlying the molecular mechanisms of flooding-tolerant responses in soybean. This result supplied a good foundation for gene function analysis in further work.
Introduction
Soybean [Glycine max (L.) Merr] provides abundant flavonoids, plant-based proteins and lipids. It is the major protein source for vegetarians. Soybean is nutritious for their isoflavones and anthocyanins belonging to flavonoid compounds1. Isoflavones, of which soybean has higher content, generally exist in many kinds of plants2. Isoflavones have been functionally linked to anti-oxidation, reduction in inflammation, inhibition of free radicals, and cancer prevention3,4,5. Anthocyanin and its main constituents, such as cyaniding-3-O-glucoside, present in soybeans can effectively inhibit lipopolysaccharide, hydrogen peroxide, and pro-inflammatory cytokines, which are a natural source of antioxidants and anti-inflammatory6,7,8. Hence, soybeans could be used to boost the nutritional content, nutraceutical products, and potential therapeutic agents for some pathological diseases.
Figure 1
The study pipelines. This pipeline consists of six steps. The first step is the GO annotations filtering. The second step is gene-wise statistic score calculation. The third step is the pathway enrichment analysis. The fourth step is the gene network construction. The fifth step is the discovery of key genes. The final step is the validation study using the qRT-PCR experiments.
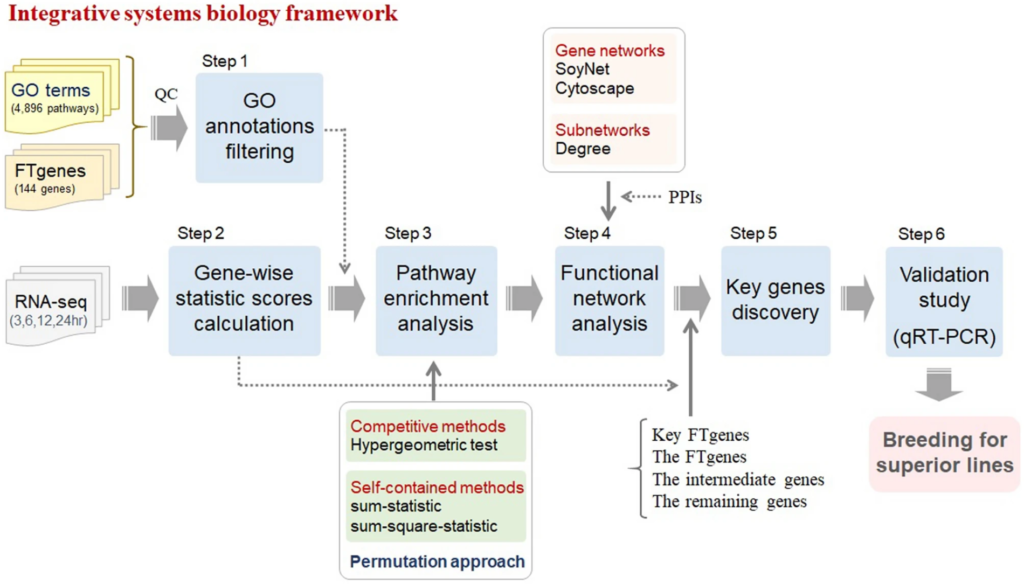
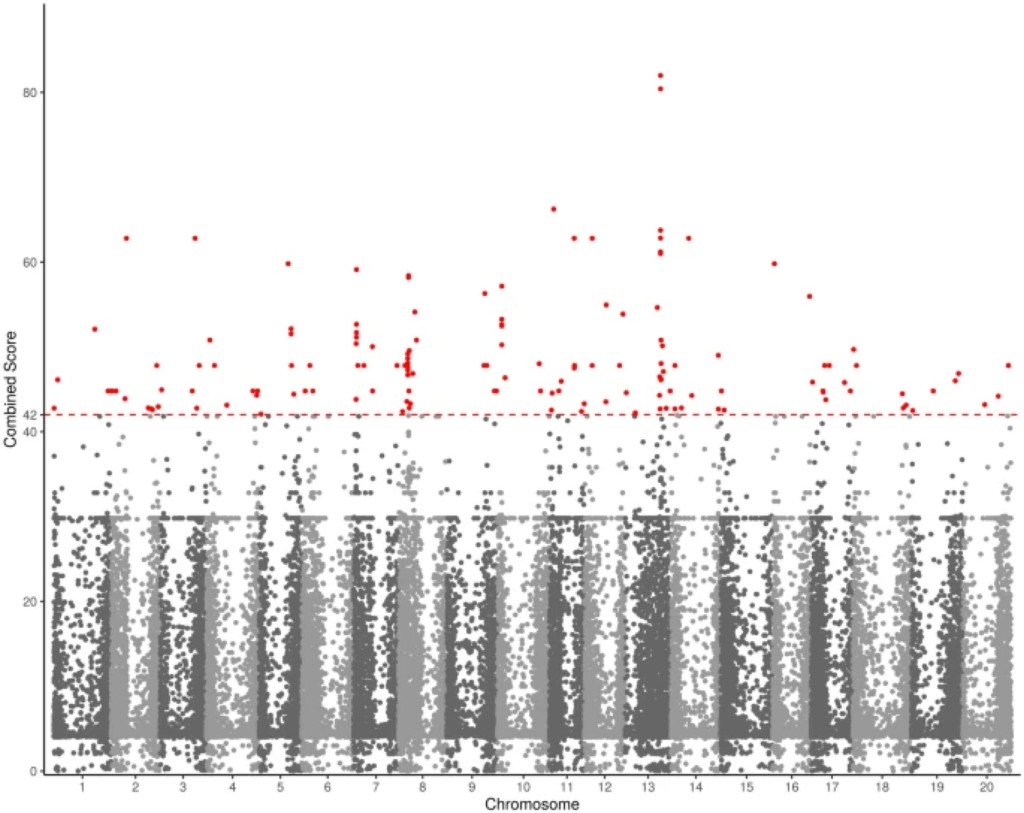
Figure 2
Manhattan plot of the flooding-tolerance genes in soybean. The dashed line is the cut-off score of 42. Dots colored in red are the FTgenes.

